The impact of systemic health upon the onset, course and recovery of a range of ocular conditions has long been known and a better understanding of this association is ongoing. Disease which influences the immune system can then have consequences upon the impact of other disease courses, and this has been brought into sharp relief recently with studies showing patients with conditions such as obesity, diabetes, even age related macular degeneration (see Optician 11.09.2020, page 18) are likely to suffer greater morbidity or mortality if they contract Covid-19.
The aim of this series is to develop the idea that eye health is linked with overall health and that there are strong associations with the overall state of the immune system which itself is very much influenced by the make-up and stability of the microbiome.
What Is the Microbiome?
A healthy body acts as host to a vast variety of microbes colonising it both externally and internally. These microorganisms, and their genetic material, are collectively known as the human microbiome. The organisms themselves are known as microbiota.1 The microbes gain a habitat and nourishment from the host and, in turn, recent research is establishing that these microbes help the host by regulating various host physiological functions. These include dietary digestion and protective immunity against pathogens.2 In short, humans require commensal microorganisms for vital functions.3
The microbiome of healthy individual consists of the following categories of microbiota:
- Bacteria
- Viruses
- Fungi
These all play a symbiotic role in maintaining homeostasis.3
Bacteria represent the most studied, abundant and diverse group, accounting for 1014 microbes,3,4 outnumbering human cells by a ratio of 10 to 1 and contributing more essential genes for survival than human cells. Whereas the human genome carries only approximately 20,000 genes, the microbiome contributes approximately eight million genes.1
The various mucosal regions (including the gastrointestinal tract) within the human body also have characteristic microbial profiles and the make-up of these is influenced by a variety of factors, ranging from pH and oxygen exposure to diet and antibiotic exposure.3
The number of bacteria at each mucosal site varies significantly. Some mucosal sites, like the gastrointestinal (GI) tract, have a bacterial genome that is more than 100 times the size of the human genome, while other surfaces are comparatively lacking in bacteria, or, paucibacterial. An example of such is the ocular surface. This translates to there being approximately 0.06 bacteria per human cell in the conjunctiva compared with approximately 12 bacteria per human cell in the buccal mucosa (in the mouth) and approximately 16 bacteria per human cell in the skin adjacent to the mouth, each with markedly different varieties of constituent microbiota.5,6 Despite the overall low number of bacteria on the ocular surface, studies have demonstrated the existence of an ocular surface microbiome in healthy individuals, which is composed of a specific community of bacteria.3
The majority of microbiota reside in the GI tract, which also accounts for almost 70% of the immune system in total. Recent research is showing that this microbiota plays an active role in some key processes including:
- Immunity homeostasis; in particular the adaptive immune system, where the microbiota interacts with the host immune system, so inducing the accumulation of several different lymphocyte populations at mucosal sites1,3,7-9
- Nutrient extraction from food
- Defend against pathogens3
Figure 1 shows the overall structure of the GIT. The microbiota is found predominantly in the colon, an area lacking in digestive enzymes. The final stage of exposure to digestive enzymes occurs in the ileum. Any macronutrients not digested pass into the colon and are further broken down through a process of fermentation by the vast array of macronutrient specific microbiota
species.10-12
 Figure 1: Overview of bacterial colony density throughout the digestive tract, factors from the proximal tract that shape small intestinal communities, and distinguishing characteristics of the small intestine and colon11
Figure 1: Overview of bacterial colony density throughout the digestive tract, factors from the proximal tract that shape small intestinal communities, and distinguishing characteristics of the small intestine and colon11
Commensal microbes colonise all exposed surfaces of the human body and outnumber our own cells by at least tenfold. The GIT is the most densely populated organ, however, and is estimated to contain around 100 trillion commensal bacteria.13
Differences and Contents of the Microbiome
The classification of living organisms into specific groups and categories, each based on a variety of common characteristics, is known as taxonomy. One common system of taxonomy categorises organisms into the following hierarchy:
• Phylum
o Class
ν Order
• Family
o Genus
ν Species
It is this system that will be referred to throughout this series.
The two dominant bacterial phyla in the healthy adult gut, accounting for almost 90% of the microbiota, are as follows:13-16
- Firmicutes; a phylum of bacteria, most of which have a Gram-positive cell wall structure and almost entirely belonging to the Clostridium genus
- Bacteroidetes; made up of three Gram negative classes of bacteria and mainly comprised of the genera Bacteroides and Prevotella
Although considerably lesser in size, the remainder consists primarily of Actinobacteria, a phylum of Gram-positive bacteria represented in the GIT by the Bifidobacterium genus.4,10,14 This is in contrast to the ocular surface microbiome which is composed of three dominant phyla,17 namely:
- Proteobacteria (accounting for 64%)
- Actinobacteria (20%)
- Firmicutes (4%)
The GIT microbiota constituents vary according to anatomical region in the intestine, and distribution is also influenced by the various local physiological activities, pH, oxygen tension, digestive flow rates, and availability of substrate.18
The Microbiome Throughout Life
The composition of the gut microbiome naturally changes throughout the course of our lives.19 At birth, the intestine is not devoid in bacteria as some might think. This is due to the presence of microbiota in the placenta and amniotic fluid.20 However, the core microbiota that colonise the GIT is shaped during early post-partum life, becoming more like that of an adult by the age of three years.15 A variety of factors impact the composition from infancy, including the following:
- Gestational age; this has a major influence upon GIT microbiota colonisation,15 and the microbiota in pre-term infants (those less than 37 weeks) differs significantly from those of full-term infants. Pre-term infants show less microbiome diversity, with lower levels of the more protective bacteria (such as Bifidobacterium, Bacteroides and Firmicutes) and an associated higher level of potentially pathogenic bacteria (from the Protobacteria phylum). This may explain the association between post-natal maturation of the GIT and systemic immunity and how it may negatively impact pre-term deliveries.21
- Mode of birth delivery; this impacts upon both the composition of the microbiota as well as the future development of the immune system.22 Vaginal delivery results in a more diverse bacterial microbiota than that found in infants delivered by caesarean section,15,22 the latter having significantly less Bifidobacteria and Bacteroides.23 This difference appears to continue into childhood.24 That the mode of delivery impacts upon the gut microbiota is of particular modern-day importance, as the prevalence of caesarean section births is increasing in the Western countries, from around 10% in the 1980s to approaching 25% in the UK today.25,26
- Mode of feeding; this plays a role in the composition of the gut microbiota, particularly in newborns.15 Breastfed infants have a richer and more diverse gut microbiota than formula fed infants,27 with a greater abundance of Bifidobacterium (Actinobacteria phyla). These are thought to strengthen metabolic and immune functioning.29 The weaning period also involves a greater amount of Bifidobacterium, Bacteroides and Clostridium (of the genus Coccoides). The differences in the microbiota between milk-fed and formula-fed becomes less marked during this weaning period.
The composition of the gut microbiome changes throughout the course of life (see figure 2). The infant microbiome shows a great inter-individual variability and relatively low diversity, but becomes more diverse and converges into an adult-like structure by the age of three years. Pregnancy is associated with an increase in Actinobacteria and Proteobacteria and increased diversity, but the gut microbiota returns to its original structure sometime after delivery. Old age (meaning over 65 years) is associated with a number of changes in the microbiota, including an increase in the abundance of Bacteroidetes.19

Figure 2: The structure of the human intestinal microbiota across the life cycle19
Bifidobacteria are important in driving metabolic and immune processes.30 A reduction in their number may explain the reduced immune system activity, incidence of malnutrition and increased prevalence of systemic disease in the elderly population.32 This highlights the usefulness in opening up the conversation about pre and probiotic supplementation for overall wellbeing in a vulnerable population.32 This argument will be further developed in future articles.
As has now been established, the composition of microbiota varies in the same individual as well between individuals. Research has found that the colon possesses an ecosystem that promotes certain clusters of microbiota organisms, called enterotypes, over others. Each individual person has a specific enterotype based upon their GIT ecosystem. Some individuals have a greater amount of Bacteroides (and classified as type I profile), others host more Prevotella (type II) and others have a greater amount of Ruminococcus (type III).14 The latter is less prevalent and can be merged into Types I and II as argued by Cheng et al in a paper that argues that, while ‘it is really pragmatic for researchers to collapse them into a few categories… Nevertheless, existing studies cannot either substantiate or deny the enterotype concept.’34
Each enterotype is based upon each individual’s dietary habits, as the microbiota derive energy from this food source. A diet rich in animal matter and carbohydrates is more associated with a type I enterotype profile, whereas a diet rich in plant-based materials is more associated with a type II profile.14 Our knowledge of enterotype based upon dietary habits may improve the understanding of the relationship between GIT microbiota and human health15,34 and this point will be further expanded upon in a later article in this series.
When the composition of the microbiome changes, a disruption in the homeostatic state occurs,35 so encouraging the growth and invasion of pathogenic species.3,36 It is to this that we will now turn our attention.
The Role of The GIT Microbiome in Maintaining Health
Human health is intricately related to how the GIT microbiome regulates the immune system. Research points to the role that GIT microbiota-derived signals play in tuning the immune cells for pro and anti-inflammatory response, thereby affecting the susceptibility to various diseases.37 The gut is surrounded by 70% of the total lymphoid tissue found in the body and is known by the acronym GALT (Gut Associated Lymphoid Tissue).2
As in many immune processes, antigen-presenting cells (APC) present antigenic material from the GIT to T-cells (specifically, T helper 17 cells usually described as Th17). If the Th17 recognises the antigen, a series of chemical processes (involving IL17 cytokines) results in an up-regulation of a pro-inflammatory response.38 The level of this response is kept in check by T-regulatory (T-reg) cells. These, effectively, ‘calm’ the immune response, limiting its potential for getting out of control. Some commensal bacteria (Firmicutes) up-regulate T-reg cells through the production of short chain fatty acids (SCFA) and reduce the potential of immune disease.3
The nature of this balancing act is summarised schematically in figure 3.

Figure 3: Immunological dysregulation is the result of dysbiosis in the microbiota.
(a) A healthy microbiota contains a balanced composition of multiple classes of bacteria. Commensals are permanent residents of this complex ecosystem and are thought to provide no benefit or detriment to the host. Symbionts are organisms with known health-promoting functions. Pathobionts are also permanent residents of the microbiota with the potential to induce pathology.
(b) During dysbiosis, there is an unnatural shift in the composition of the microbiota whereby either the numbers of symbionts are reduced and/or pathobionts are increased. The various causes for this are not entirely clear, but are likely to include recent societal advances in developed countries. The result is non-specific inflammation which may predispose certain genetically susceptible people to inflammatory disease. Pathogens are opportunistic organisms that cause rare and acute inflammation.9
The Gut-Microbiota Axes
The influence of the microbiota goes beyond the GI tract.4,15 Research demonstrates the existence of bi-directional communication between the gut microbiota and other sites. This leads us to the concept of many gut-‘target organ’ axes, each having an influence upon a variety of systemic effects.39
Of relevance to the eye care professional, the gut-brain axis has implications for central nervous system disorders. Of particular topicality at present is the prospect of a gut-lung axis. Let us consider the latter first.
Gut-Lung Axis and COVID
In this Covid-19 world, it is of particular interest that a gut-lung axis exists and represents a means of bi-directional communication which may impact upon the lung and result in inflammation, as well as reciprocally impacting upon the gut microbiota.40
Recent research is supporting the existence of a potential route of viral entry via receptors common in the lungs and gut.2 Interestingly, recent research has also shown a consistent alteration in the gut microbiota of a small number of hospitalised Covid-19 patients and a potential link to disease severity has been postulated.41 The potential implication is that targeting of the gut microbiome may represent a novel therapeutic approach to improve clinical outcomes of Covid-19 in the future.42-44
Gut-Brain Axis
Of interest for eye care clinicians is the gut-brain axis13,45 as with the concept are implications that the GIT microbiota affects diseases in the central nervous system (CNS). This bi-directional communication involves the brain modulating the GIT through the regulation of motility, secretion, absorption and blood flow. There is also the implication of the GIT affecting brain function.39,47 A number of pathways have been proposed to explain this bidirectional communication:
- Activation of the vagus nerve by gut microbiota.47 The vagus (cranial nerve X) runs from brainstem to GIT, regulating several vital functions. These include bronchial constriction, heart rate and gut motility.48 The vagus appears to differentiate between non-pathogenic and potentially pathogenic bacteria, even in the absence of overt inflammation. Certain vagal signals from the gut can instigate anti-inflammatory afferent signals to the brain. These signals, in turn, produce an efferent response, whereby mediators are released. These interact with immune cells and reduce inflammation.49 This immunomodulatory role of the vagus nerve may also have consequences for modulation of brain function and mood.47
- Direct action upon the immune system.50 The immune cells from the GALT (such as T-cells, macrophages, and dendritic cells) can cross the blood-brain barrier (BBB) and influence neurons and glia in the brain.52 Therefore, gut microbiota can also modulate the activity of resident immune cells, in addition to their effects on neuronal cells in the CNS.52 Systemic circulation of immune factors, cytokines and chemokines influences the brain via the vagus nerve53 and drainage of the lymphatic system into the brain provide more decisive evidence of direct cytokine entry into the brain, enabling interaction with neural tissues.54
- Gut microbiota impact the amino acid tryptophan. Tryptophan is essential in the synthesis of a variety of mediators, including the neurotransmitter serotonin which plays a critical role in CNS regulation.55-57 That the diversity and stability of the GIT microbiota varies with age may account for serotonin-related health problems in the elderly.57
- Impact upon the production of metabolites. An influence upon metabolites such as short-chain fatty acids (SCFAs) that possess neuroactive properties, may influence CNS health.58
GIT microbiota are also capable of producing a variety of neurotransmitters and neuroactive compounds themselves,59 as summarised in table 1.
 Table 1: Microbiota and their neurochemical products
Table 1: Microbiota and their neurochemical products
The Role of Short Chain Fatty Acids
Diet influences the composition of the GIT microbiota with specific changes to the major macronutrient contained in the diet. The GIT plays an important role in the metabolising of indigestible macronutrients and this results in the production of bioactive compounds, including short chain fatty acids (SCFA).4,10
SFCAs are derived from dietary fibre60 and are used both for energy production and act as a signalling molecule in various tissues.16 Humans lack the enzymes to breakdown the majority of dietary fibre (typically polysaccharide carbohydrate, such as cellulose), which instead passes undigested into the large intestine (see figure 1). Here, fermentation takes place, mainly in the proximal colon, by virtue of the gut microbiota.61 This fermentation process yields a variety of metabolites of which SCFAs form the major group, accounting for approximately 10% of our caloric requirements.62 These SCFAs play an important role in a variety of systemic conditions including ulcerative colitis, Crohn’s disease and colorectal cancer.63,64
SCFAs are saturated organic compounds consisting of one to six carbon chains, the most abundant being acetate (C2), priopionate (C3) and butyrate (C4).4,65 Although the quantity of SCFAs reduces from the proximal to the distal colon, the concentration remains at a stable proportion (60:20:20) throughout.66 The fermentation by-products from the microbiota vary depending upon their phylum. The Bacteroidetes produce acetate and propionate, whereas the Firmicutes have butyrate as the primary metabolic end product.67
The production of SCFAs from the bacterial fermentation process involves a number of pathways, each requiring the microbiota to work as a community as well as having a symbiotic relationship with the host.4
Changes in the amount and type of fibre ingested will have a knock-on effect upon the type and quantity of SCFAs. This impacts upon the pH of the colon, which then affects the composition of the microbiota, which in turn affects further SCFA production.4
A typical Western diet, with a greater amount of meat and reduced vegetable intake, results in increased advanced glycation end (AGE) products and lower production of SCFAs. The net increase in AGE products induces changes in the gut microbiota that favour greater insulin resistance and are considered a risk factor for metabolic diseases (such as obesity, type 2 diabetes mellitus,68 and cardiovascular disease69) compared to a more vegetable diet. A diet richer in vegetables, on the other hand, produces more SCFAs, so improving insulin sensitivity and regulating its secretion and is thought to play a beneficial role in reducing the risks of type 2 diabetes.70,71
As already mentioned, there is greater production of SCFAs in the proximal colon. This results in a lower pH which favours more butyrate-producing bacteria (from the Firmicutes phylum).72 The lower pH has the benefit of reducing the chance of an overgrowth of pH-sensitive pathogenic bacteria, such as Clostridium spp.4 In contrast, fermentable dietary fibre is less abundant in the distal colon, resulting in lower SCFA production and hence a relative raising of pH, so favouring the acetate and propionate-producing bacteria (from the Bacteroidetes phylum).72
Once produced, the SCFAs have the ability to exert influence systemically. The majority are absorbed by colonocytes and also bind with and activate certain receptors lining the lumen of the colon (G Protein-coupled receptors or GPCRs), impacting intestinal mucosal immunity as well as barrier integrity.16,39 A smaller fraction of SCFAs enter the systemic circulation73 and exert a beneficial influence on fat tissue activation and effecting a reduction in appetite,74 regulation of mitochondrial liver function, sleep modulation75 as well as increasing insulin secretion.76
SCFAs cross the BBB and have been detected in the cerebrospinal fluid,77 the brain16,79 and, most recently have been implicated in the regulation of epigenetic gene expression.79 Figure 4 offers a schematic representation of how SCFAs influence gut-brain communication.
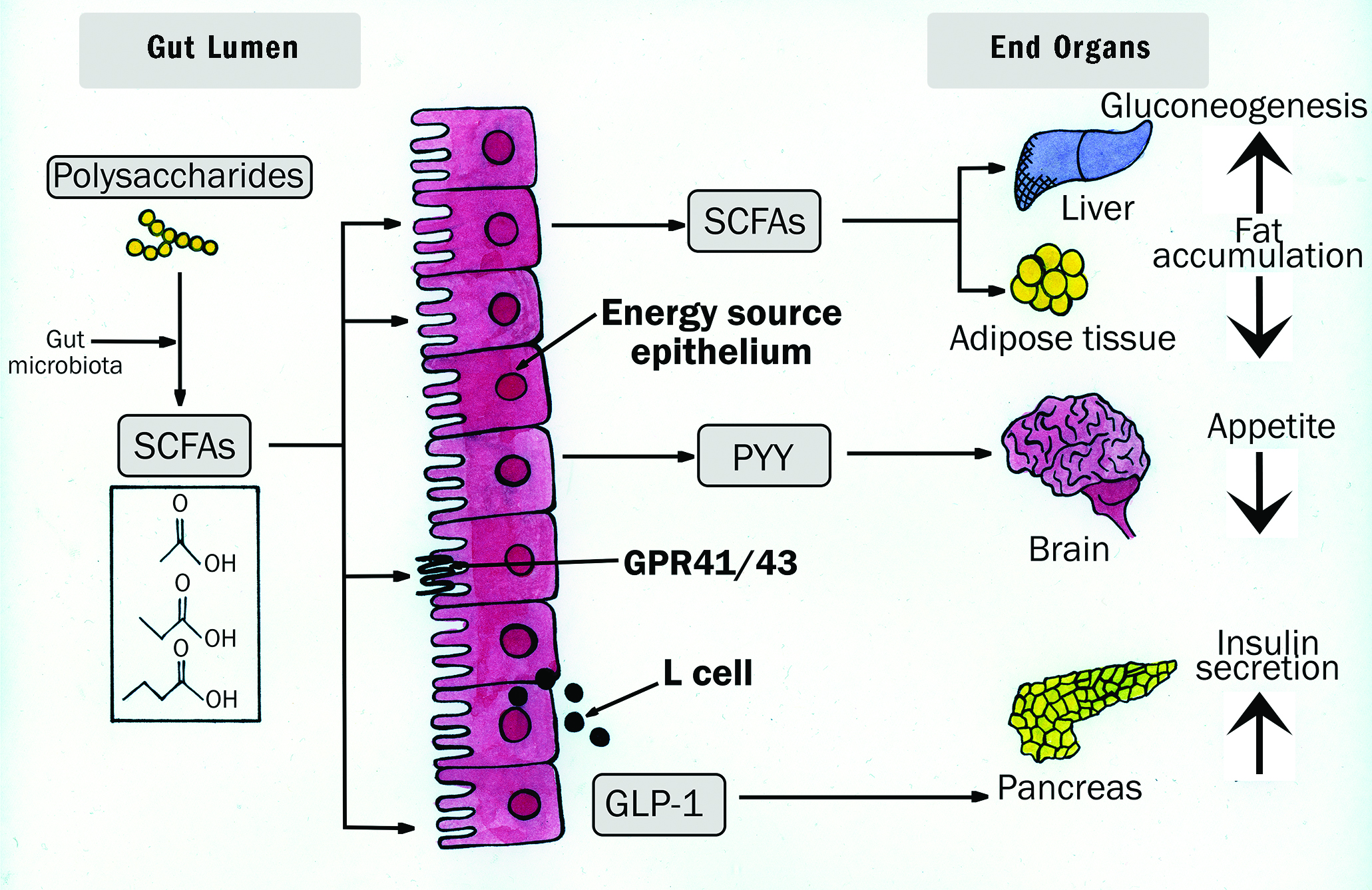 Figure 4: Schematic representation of the nature of SCFA influence upon the gut-brain axis16
Figure 4: Schematic representation of the nature of SCFA influence upon the gut-brain axis16
Dysbiosis
When the composition of the microbiome alters, homeostasis can be lost. This is termed dysbiosis and it can increase the likelihood of an overgrowth or opportunistic invasion of pathogenic bacteria.
Dysbiosis may be caused by a number of factors,3 including:
- Diet
- Use of antibiotics
- Infection
- Ageing
Specific properties of antibiotics, such as antimicrobial effects or mode of action, are partially responsible for the shifts in bacterial composition during antibiotic therapy.15 Broad-spectrum antibiotics, for example, lead to an imbalance between Firmicutes and Bacteroidetes. An example of this is Clostridium difficile infection following antibiotic use. Clostridium difficile forms part of a healthy gut microbiome, playing a role in the production of SCFA.3,80 However, the use of oral antibiotics can alter the
protective mucous layer lining the GIT leading to a loss of the integrity of the epithelial barrier, changes of the mucosal immune system and impaired absorption of nutrients.80 This ultimately promotes bacterial overgrowth and fuels a C. difficile inflammatory process, resulting in gastrointestinal disorders (irritable bowel syndrome) and even colorectal cancer.
Theories suggest that intestinal dysbiosis can disrupt intestinal barrier function and result in the leakage of bacteria or their antigens into the blood stream or lymph system, increasing the potential for the dysregulation of the extraintestinal adaptive immune response.65
Recent studies have suggested that the GIT microbiota dysbiosis may be closely linked to a range of host disorders, including the following:
- Obesity82
- Type 2 diabetes82,83
- Inflammatory conditions; these include inflammatory bowel diseases (including ulcerative colitis and Crohn’s disease)16
This GIT dysbiosis also appears to have an association with other inflammatory diseases beyond the GIT,65 including rheumatoid arthritis,84 ankylosing spondylitis85 and multiple sclerosis.86 Other associated neurological disorders of interest, and that have been associated with alterations in the gut microbiota,70 include the following:
- Stress
- Autism
- Depression
- Parkinson’s disease
- Alzheimer’s disease
And Finally… the Eye
As there also appears to be a gut-retina axis,87 GIT dysbiosis has been implicated in altering the immune system and so affecting a variety of ocular conditions (figure 5).3 These include:
- Anterior uveitis
- Posterior uveitis
- Diabetic retinopathy
- Glaucoma
- Age-related macular degeneration
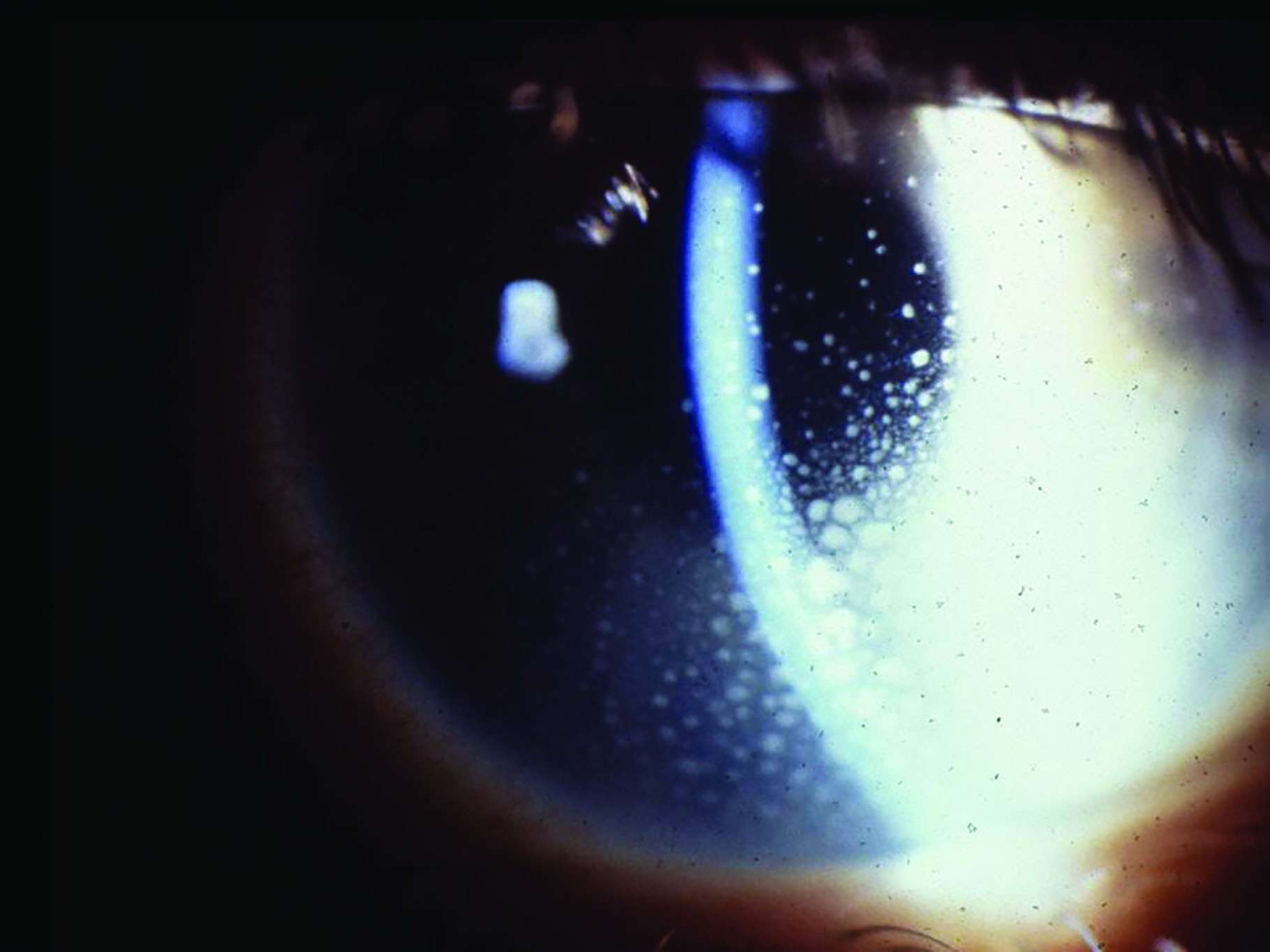 Figure 5: Anterior uveitis
Figure 5: Anterior uveitis
 Figure 5: Posterior uveitis
Figure 5: Posterior uveitis
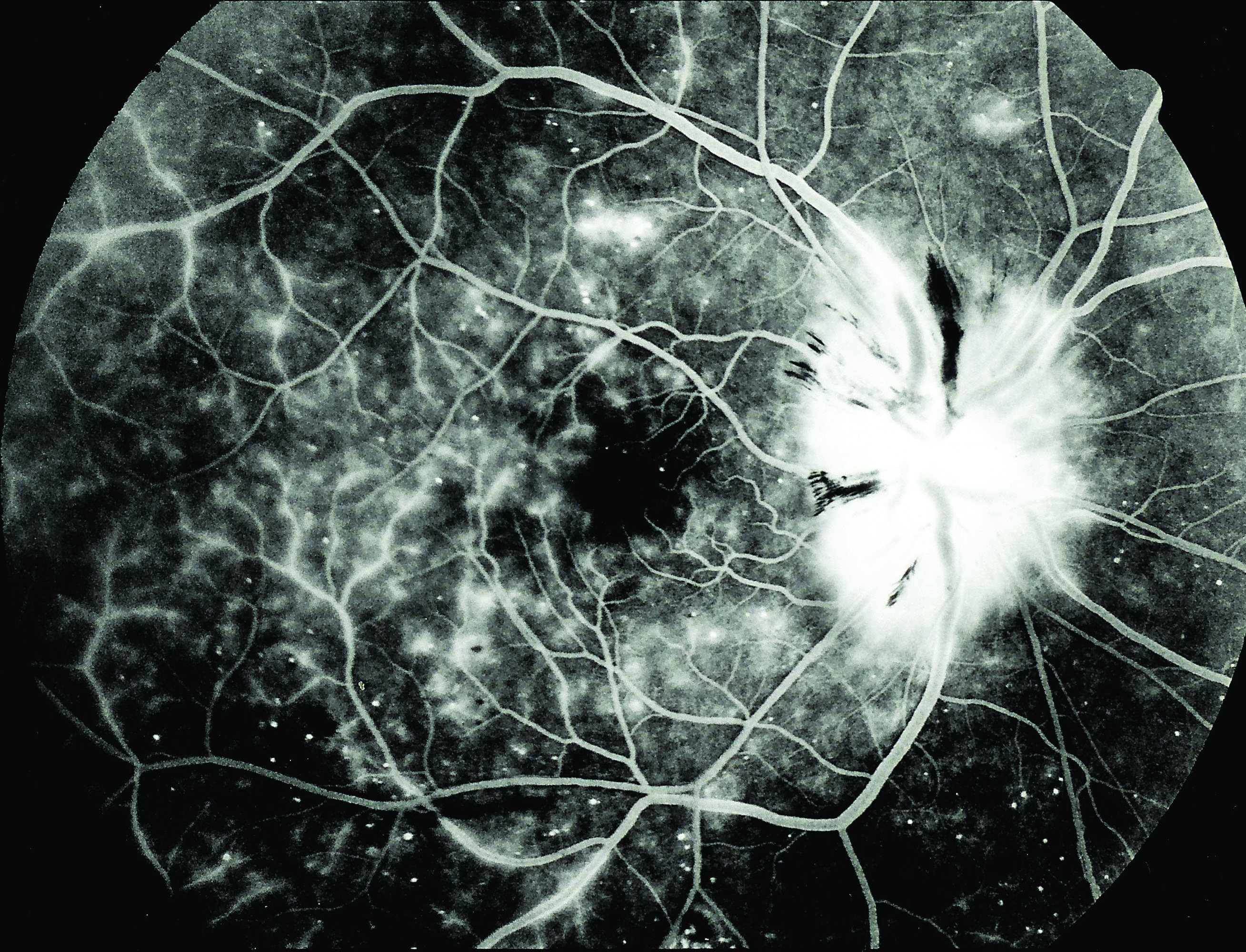 Figure 5: Diabetic retinopathy
Figure 5: Diabetic retinopathy
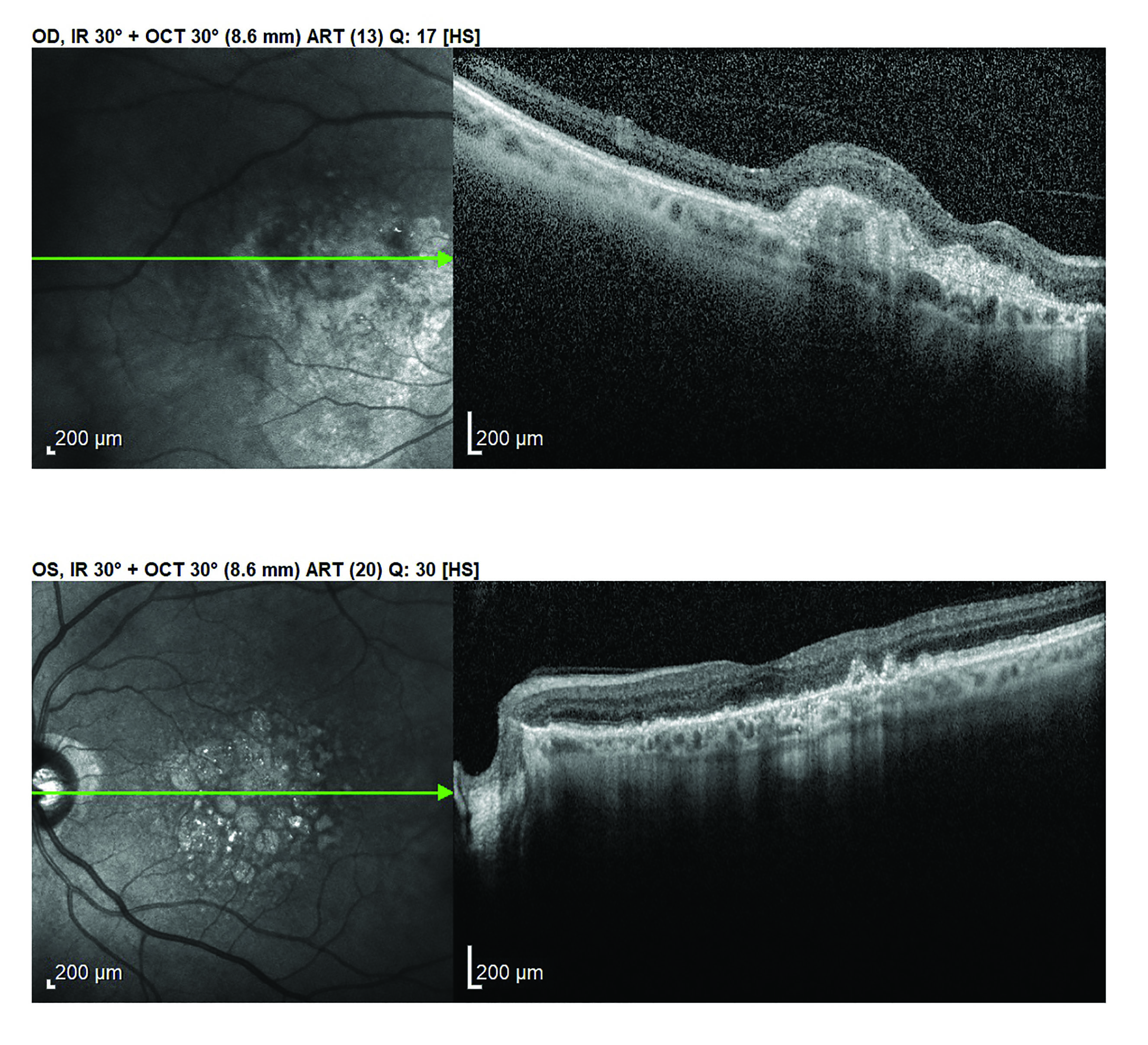 Figure 5: Age-related macular degeneration
Figure 5: Age-related macular degeneration
Subsequent articles will concentrate on the role inflammation plays in age-related macular degeneration and its link with the GIT microbiome. The final article will consider how a more holistic approach may influence our patient assessment and management in daily clinical practice.
- Dr Rohit Narayan is a therapeutic optometrist and Clinical Scientist at Aston University.
Acknowledgments
- Figures 1 to 4 courtesy of Kitty Harvey.
References
- Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, Warner BB, Tarr PI, Wang D, Holtz LR. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. 2015;21:1228–34
- Dhar, D and Mohanty, A. Gut microbiota and Covid-19-possible link and implications. Virus Research, 2020, p.198018
- Cavuoto, KM, Banerjee, S and Galor, A. Relationship between the microbiome and ocular health. The Ocular Surface, 2019.
- den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54(9):2325-2340
- Doan, T, Akileswaran, L, Andersen, D, Johnson, B, Ko, N, Shrestha, A, Shestopalov, V, Lee, CS, Lee, AY and Van Gelder, RN, 2016. Paucibacterial microbiome and resident DNA virome of the healthy conjunctiva. Investigative Ophthalmology & Visual Science, 57(13), pp.5116-5126
- Ozkan, J and Willcox, MD, 2019. The ocular microbiome: molecular characterisation of a unique and low microbial environment. Current Eye Research, 44(7), pp.685-694
- Atarashi, K, Tanoue, T, Shima, T, Imaoka, A, Kuwahara, T, Momose, Y, Cheng, G, Yamasaki, S, Saito, T, Ohba, Y and Taniguchi, T, 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science, 331(6015), pp.337-341
- Collins, SM, Surette, M and Bercik, P, 2012. The interplay between the intestinal microbiota and the brain. Nature Reviews Microbiology, 10(11), pp.735-742
- Round, JL and Mazmanian, SK, 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nature reviews immunology, 9(5), pp.313-323
- Alou, MT, Lagier, JC and Raoult, D, 2016. Diet influence on the gut microbiota and dysbiosis related to nutritional disorders. Human Microbiome Journal, 1, pp.3-11
- Kastl Jr, AJ, Terry, NA, Wu, GD and Albenberg, LG. The structure and function of the human small intestinal microbiota: current understanding and future directions. Cellular and Molecular Gastroenterology and Hepatology, 2020, 9(1), pp.33-45
- Maukonen, J and Saarela, M. Human gut microbiota: does diet matter? Proceedings of the Nutrition Society. Cambridge University Press, 2015, 74(1), pp. 23–36
- Caspi, RR and Horai, R. Microbiome and autoimmune uveitis. Frontiers in Immunology, 2019, 10, p.232
- Arumugam, M, Raes, J, Pelletier, E, Le Paslier, D, Yamada, T, Mende, DR, Fernandes, GR, Tap, J, Bruls, T, Batto, JM and Bertalan, M. Erratum: Enterotypes of the human gut microbiome (Nature (2011) 473 (174-180)). Nature, 474(7353)
- Rinninella, E, Raoul, P, Cintoni, M, Franceschi, F, Miggiano, GAD, Gasbarrini, A and Mele, MC. What is the healthy gut microbiota composition? a changing ecosystem across age, environment, diet, and diseases. Microorganisms, 2019, 7(1), p.14
- Shimizu, H, Ohue-Kitano, R and Kimura, I. Regulation of host energy metabolism by gut microbiota-derived short-chain fatty acids. Glycative Stress Research, 2019, 6(3), pp.181-191
- Dong, Q, Brulc, JM, Iovieno, A, Bates, B, Garoutte, A, Miller, D, Revanna, KV, Gao, X, Antonopoulos, DA, Slepak, VZ and Shestopalov, VI. Diversity of bacteria at healthy human conjunctiva. Investigative Ophthalmology & Visual Science, 2011 52(8), pp.5408-5413
- Flint, HJ, Scott, KP, Louis, P and Duncan, SH. The role of the gut microbiota in nutrition and health. Nature Reviews Gastroenterology & Hepatology, 2012, 9(10), p.577
- Kostic, AD, Howitt, MR and Garrett, WS. Exploring host–microbiota interactions in animal models and humans. Genes & Development, 2013, 27(7), pp.701-718
- Collado, MC, Rautava, S, Aakko, J, Isolauri, E and Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Scientific Reports, 2016, 6, p.23129
- Ren, S, Hui, Y, Obelitz-Ryom, K, Brandt, AB, Kot, W, Nielsen, DS, Thymann, T, Sangild, PT and Nguyen, DN. Neonatal gut and immune maturation is determined more by postnatal age than by postconceptional age in moderately preterm pigs. American Journal of Physiology-Gastrointestinal and Liver Physiology, 2018, 315(5), pp. G855-G867
- Biasucci, G, Benenati, B, Morelli, L, Bessi, E and Boehm, G. Cesarean delivery may affect the early biodiversity of intestinal bacteria. The Journal of Nutrition, 2008, 138(9), pp.1796S-1800S.
- Biasucci, G, Rubini, M, Riboni, S, Morelli, L, Bessi, E and Retetangos, C. Mode of delivery affects the bacterial community in the newborn gut. Early Human Development, 2010, 86(1), pp.13-15
- Salminen, S, Gibson, GR, McCartney, AL. and Isolauri, E. Influence of mode of delivery on gut microbiota composition in seven-year-old children. Gut, 2004, 53(9), pp.1388-1389
- Bragg, F, Cromwell, DA, Edozien, L., Gurol-Urganci, I, Mahmood, TA, Templeton, A and van der Meulen, JH. Variation in rates of caesarean section among English NHS trusts after accounting for maternal and clinical risk: cross sectional study. BMJ, 2010, 341, p.c5065
- Parlimentary Office of Science and Technology: Caesarean Sections. www.parliament.uk/documents/post/pn184.pdf
- Penders, J, Thijs, C, Vink, C, Stelma, FF, Snijders, B, Kummeling, I, van den Brandt, PA and Stobberingh, EE. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics, 2006, 118(2), pp.511-521
- Clarke S. F., Murphy E. F., Nilaweera K., Ross P. R., Shanahan F., O’Toole P. W., et al. The gut microbiota and its relationship to diet and obesity: new insights. Gut Microbiology, 2012, 3, 186–202
- Fallani, M., Amarri, S., Uusijarvi, A., Adam, R., Khanna, S., Aguilera, M., Gil, A., Vieites, J.M., Norin, E., Young, D. and Scott, J.A. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology, 2011, 157(5), pp.1385-1392
- Arboleya, S., Watkins, C., Stanton, C. and Ross, R.P. Gut bifidobacteria populations in human health and aging. Frontiers in microbiology, 2016, 7, p.1204
- Odamaki, T., Kato, K., Swugahara, H., Hashikura, N., Takahashi, S., Xiao, J.Z., Abe, F. and Osawa, R. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC microbiology, 2016, 16(1), pp.1-12
- Guigoz, Y., Doré, J. and Schiffrin, E.J. The inflammatory status of old age can be nurtured from the intestinal environment. Current Opinion in Clinical Nutrition & Metabolic Care, 2008, 11(1), pp.13-20
- Cheng, Mingyue, and Kang Ning. Stereotypes About Enterotype: the Old and New Ideas. Genomics, Proteomics & Bioinformatics, 2019, vol. 17,1, 4-12
- Christensen, L., Roager, H.M., Astrup, A. and Hjorth, M.F. Microbial enterotypes in personalized nutrition and obesity management. The American Journal of Clinical Nutrition, 2018, 108(4), pp.645-651
- Alonso, V.R. and Guarner, F. Linking the gut microbiota to human health. British Journal of Nutrition, 2013, 109(S2), pp. S21-S26
- Gritz, E.C. and Bhandari, V. The human neonatal gut microbiome: a brief review. Frontiers in Pediatrics, 2015, 3, p.17
- Negi S, Das DK, Pahari S, Nadeem S, Agrewala JN. Potential Role of Gut Microbiota in Induction and Regulation of Innate Immune Memory. Frontiers of Immunology, 2019;10:2441
- Khader, S.A., Gaffen, S.L. and Kolls, J.K. Th17 cells at the crossroads of innate and adaptive immunity against infectious diseases at the mucosa. Mucosal Immunology, 2009, 2(5), pp.403-411
- Silva, Y.P., Bernardi, A. and Frozza, R.L. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Frontiers in Endocrinology, 2020, 11, p.25
- Dumas, A., Bernard, L., Poquet, Y., Lugo‐Villarino, G., & Neyrolles, O. The role of the lung microbiota and the gut–lung axis in respiratory infectious diseases. Cellular Microbiology, 2018, 20(12), e12966
- Zuo, T., Zhang, F., Lui, G.C., Yeoh, Y.K., Li, A.Y., Zhan, H., Wan, Y., Chung, A., Cheung, C.P., Chen, N. and Lai, C.K.,. Alterations in Gut Microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology, 2020, S0016-5085, (20), pp 34701-6
- Gu, S., Chen, Y., Wu, Z., Chen, Y., Gao, H., Lv, L., Guo, F., Zhang, X., Luo, R., Huang, C. and Lu, H., 2020. Alterations of the Gut Microbiota in Patients with COVID-19 or H1N1 Influenza. Clinical Infectious Diseases, 2020, ciaa709, https://doi.org/10.1093/cid/ciaa709
- Mak, Joyce W.Y., Chan, Francis K.L., Ng, Siew C. Probiotics and Covid-19: one size does not fit all. The Lancet Gastroenterology and Hepatology, 2020; Vol. 5, No. 7
- Marcialis, M.A., Bardanzellu, F. and Fanos, V., 2020. Microbiota and Covid-19. Which came first, the chicken or the egg? Clinical Infectious Diseases, ciaa965, https://doi.org/10.1093/cid/ciaa965
- Carabotti, M., Scirocco, A., Maselli, M.A. and Severi, C. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Annals of gastroenterology: quarterly publication of the Hellenic Society of Gastroenterology, 2015, 28(2), p.203
- Kim, C.H. Microbiota or short-chain fatty acids: which regulates diabetes?. Cellular & Molecular Immunology, 2018, 15(2), pp.88-91
- Forsythe, P., Bienenstock, J. and Kunze, W.A. Vagal pathways for microbiome-brain-gut axis communication. In Microbial endocrinology: the microbiota-gut-brain axis in health and disease (pp. 115-133). 2014, Springer, New York, NY
- Bercik, P., Park, A.J., Sinclair, D., Khoshdel, A., Lu, J., Huang, X., Deng, Y., Blennerhassett, P.A., Fahnestock, M., Moine, D. and Berger, B. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut–brain communication. Neurogastroenterology & Motility, 2011, 23(12), pp.1132-1139
- Bonaz, B., Bazin, T. and Pellissier,S. The vagus nerve at the interface of the microbiota-gut-brain axis. Frontiers in Neuroscience, 2018, 12, p.49
- Bengmark, S. Gut microbiota, immune development and function. Pharmacological Research, 2013, 69(1), pp.87-113
- Diamond, B., Huerta, P.T., Tracey, K. and Volpe, B.T. It takes guts to grow a brain: increasing evidence of the important role of the intestinal microflora in neuro‐and immune‐modulatory functions during development and adulthood. Bioessays, 2011, 33(8), pp.588-591
- Berer K, Krishnamoorthy G. Commensal gut flora and brain autoimmunity: a love or hate affair? Acta Neuropathology, 2012;123(5):639-651
- Hosoi, T., Okuma, Y. and Nomura, Y. The mechanisms of immune-to-brain communication in inflammation as a drug target. Current Drug Targets-Inflammation & Allergy, 2002, 1(3), pp.257-262
- Sun, B.L., Wang, L.H., Yang, T., Sun, J.Y., Mao, L.L., Yang, M.F., Yuan, H., Colvin, R.A. and Yang, X.Y. Lymphatic drainage system of the brain: A novel target for intervention of neurological diseases. Progress in Neurobiology, 2018, 163, pp.118-143
- Agus, A., Planchais, J. and Sokol, H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host & Microbe, 2018, 23(6), pp.716-724
- Höglund, E., Øverli, Ø. and Winberg, S. Tryptophan metabolic pathways and brain serotonergic activity: a comparative review. Frontiers in Endocrinology, 2019, 10, p.158
- O’Mahony, S.M., Clarke, G., Borre, Y.E., Dinan, T.G. and Cryan, J.F. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behavioural Brain Research, 2015, 277, pp.32-48
- Dalile, B., Van Oudenhove, L., Vervliet, B. and Verbeke, K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nature Reviews Gastroenterology & Hepatology, 2019;16(8):461-478
- Lyte, M. ed., 2010. Microbial endocrinology: interkingdom signalling in infectious disease and health. New York, NY: Springer
- Kimura, I., Ichimura, A., Ohue-Kitano, R. and Igarashi, M. Free fatty acid receptors in health and disease. Physiological reviews, 2020, 100(1), pp.171-210
- Morrison, D.J. and Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut microbes, 2016, 7(3), pp.189-200
- Bergman, E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiological reviews, 1990, 70(2), pp.567-590
- Breuer, R.I., Buto, S.K., Christ, M.L., Bean, J., Vernia, P., Paoluzi, P., Di Paolo, M.C. and Caprilli, R. Rectal irrigation with short-chain fatty acids for distal ulcerative colitis. Digestive Diseases and Sciences, 1991, 36(2), pp.185-187
- Tang, Y., Chen, Y., Jiang, H., Robbins, G.T. and Nie, D. G‐protein‐coupled receptor for short‐chain fatty acids suppresses colon cancer. International Journal of Cancer, 2011, 128(4), pp.847-856
- Kim, D., Zeng, M.Y. and Núñez, G. The interplay between host immune cells and gut microbiota in chronic inflammatory diseases. Experimental & molecular medicine, 2017, 49(5), pp.e339-e339
- Topping, D.L. and Clifton, P.M. Short-chain fatty acids and human colonic function: roles of resistant starch and non-starch polysaccharides. Physiological reviews, 2001, 81:1031-1064
- Macfarlane, S. andw Macfarlane, G.T. Regulation of short-chain fatty acid production. Proceedings of the Nutrition Society, 2003, 62(1), pp.67-72
- Yamagishi, S.I. Role of advanced glycation end products (AGEs) in osteoporosis in diabetes. Current Drug Targets, 2011, 12(14), pp.2096-2102
- Hegab, Z., Gibbons, S., Neyses, L. and Mamas, M.A. Role of advanced glycation end products in cardiovascular disease. World Journal of Cardiology, 2012, 4(4), p.90
- Kim, K. N., Yao, Y., & Ju, S. Y. Short Chain Fatty Acids and Fecal Microbiota Abundance in Humans with Obesity: A Systematic Review and Meta-Analysis. Nutrients, 2018, 11(10), 2512
- Tang, C., Ahmed, K., Gille, A., Lu, S., Gröne, H.J., Tunaru, S. and Offermanns, S. Loss of FFA2 and FFA3 increases insulin secretion and improves glucose tolerance in type 2 diabetes. Nature medicine, 2015, 21(2), pp.173-177
- Walker, A.W., Duncan, S.H., Leitch, E.C.M., Child, M.W. and Flint, H.J. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Applied and environmental microbiology, 71(7), 2005, pp.3692-3700
- Cummings, J., Pomare, E.W., Branch, W.J., Naylor, C.P. and Macfarlane, G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut, 1987, 28(10), pp.1221-1227
- Li, Z., Yi, C.X., Katiraei, S., Kooijman, S., Zhou, E., Chung, C.K., Gao, Y., van den Heuvel, J.K., Meijer, O.C., Berbée, J.F. and Heijink, M. Butyrate reduces appetite and activates brown adipose tissue via the gut-brain neural circuit. Gut, 2018, 67(7), pp.1269-1279
- Szentirmai, É., Millican, N.S., Massie, A.R. and Kapás, L. Butyrate, a metabolite of intestinal bacteria, enhances sleep. Scientific Reports, 2019, 9(1), pp.1-9
- Mollica, M.P., Raso, G.M., Cavaliere, G., Trinchese, G., De Filippo, C.,
